New visualization function tutorial¶
[1]:
import healpy as hp
from healpy.newvisufunc import projview, newprojplot
[2]:
import matplotlib.pyplot as plt
import numpy as np
[3]:
m = hp.read_map(
"../test/data/wmap_band_iqumap_r9_7yr_W_v4_udgraded32_masked_smoothed10deg_fortran.fits"
);
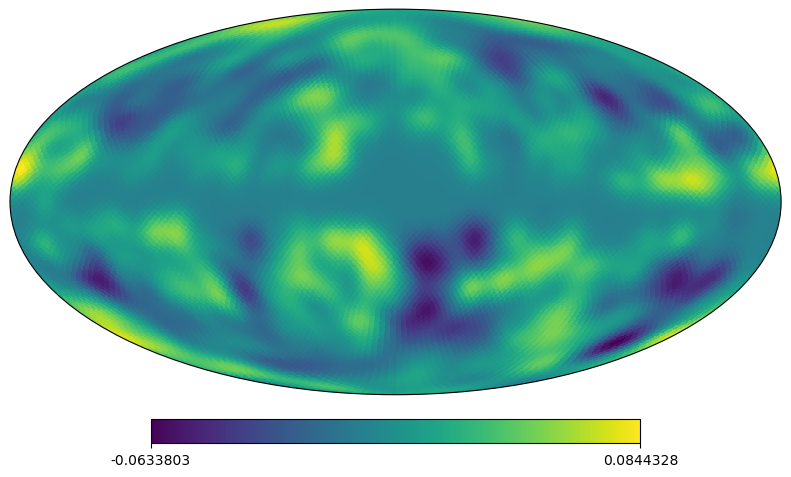
[4]:
# classic healpy mollweide projections plot
# other matplotlib projections are possible, such as: "aitoff","hammer",lambert"
projview(m, coord=["G"], flip="astro", projection_type="mollweide");
[5]:
# classic healpy mollweide projections plot with graticule
projview(
m, coord=["G"], graticule=True, graticule_labels=True, projection_type="mollweide"
);
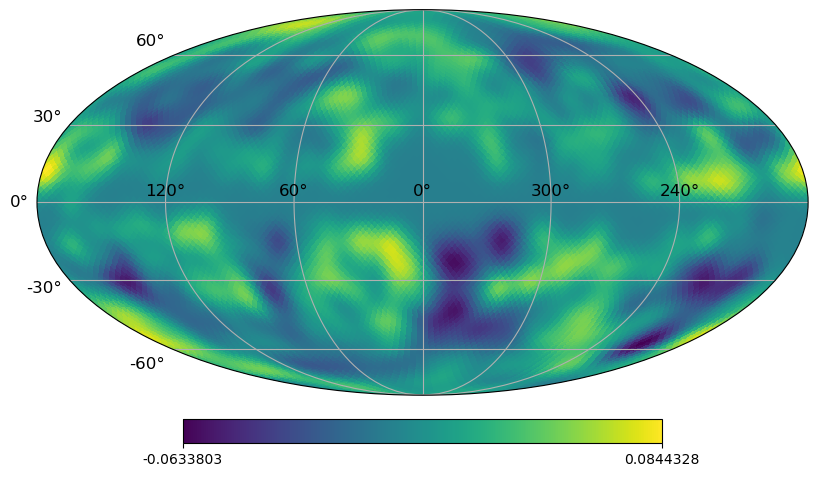
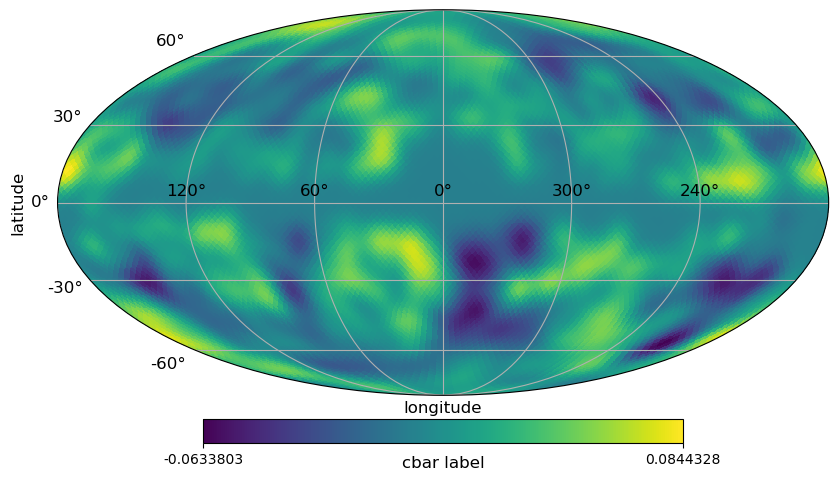
[6]:
# classic healpy mollweide projections plot with graticule and axis labels
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="horizontal",
projection_type="mollweide",
);
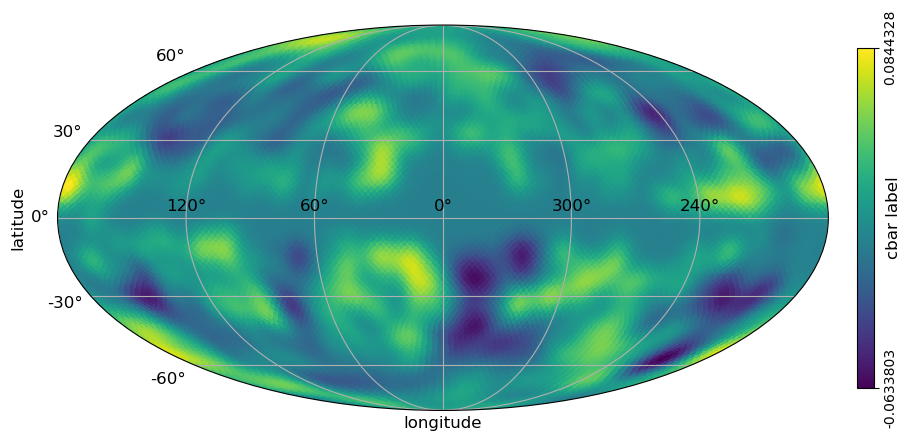
[7]:
# classic healpy mollweide projections plot with graticule and axis labels and vertical color bar
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
projection_type="mollweide",
)
[7]:
<matplotlib.collections.QuadMesh at 0x7fb384838ca0>
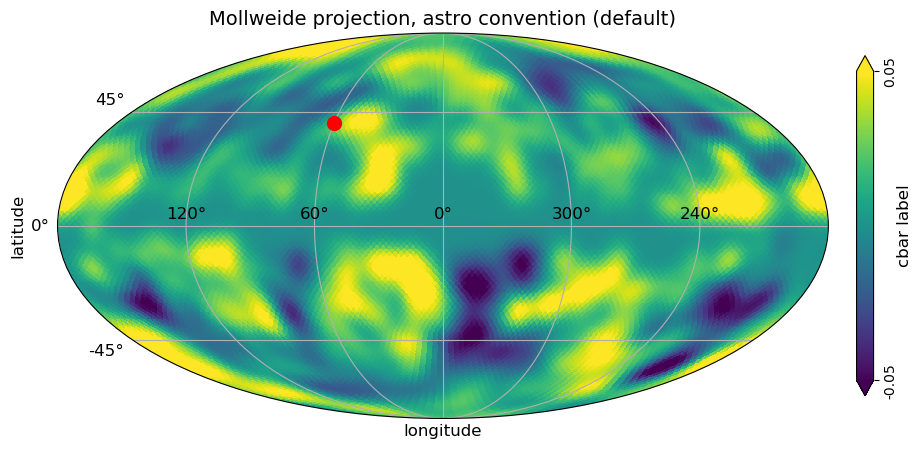
[8]:
# classic healpy mollweide projections plot with graticule and axis labels and vertical color bar, when min and max is set
# to be different than the min, max of the data, the colorbar is extended
# with title
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=45,
projection_type="mollweide",
title="Mollweide projection, astro convention (default)",
);
newprojplot(theta=np.radians(50), phi=np.radians(60), marker="o", color="r", markersize=10);
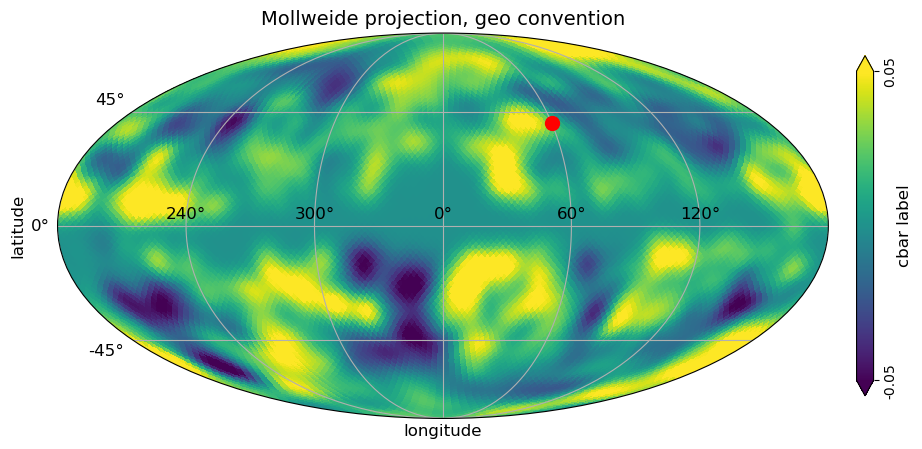
[9]:
# classic healpy mollweide projections plot with graticule and axis labels and vertical color bar, when min and max is set
# to be different than the min, max of the data, the colorbar is extended
# with title, GEO convention for longitude
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=45,
projection_type="mollweide",
title="Mollweide projection, geo convention",
flip="geo",
phi_convention="clockwise"
);
# newprojplot knows about the flip in plotting longitude
newprojplot(theta=np.radians(50), phi=np.radians(60), marker="o", color="r", markersize=10);
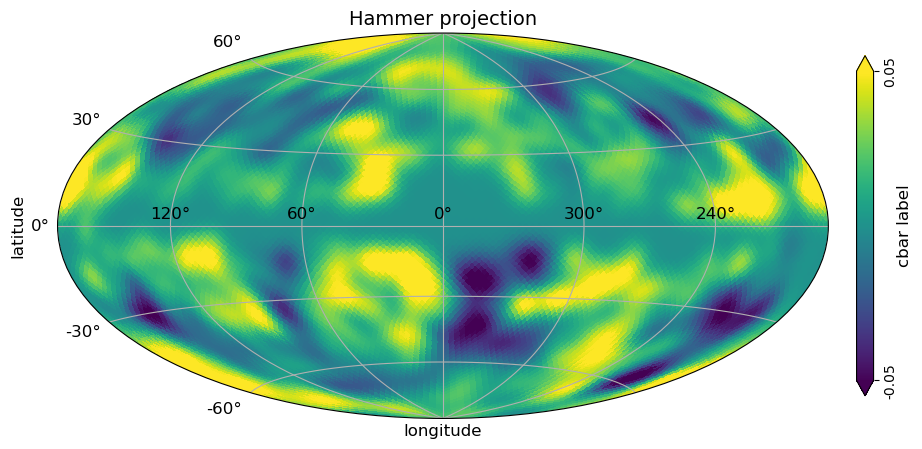
[10]:
# hammer view plot with graticule and axis labels and vertical color bar, when min and max is set
# to be different than the min, max of the data, the colorbar is extended
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="hammer",
title="Hammer projection",
);
[11]:
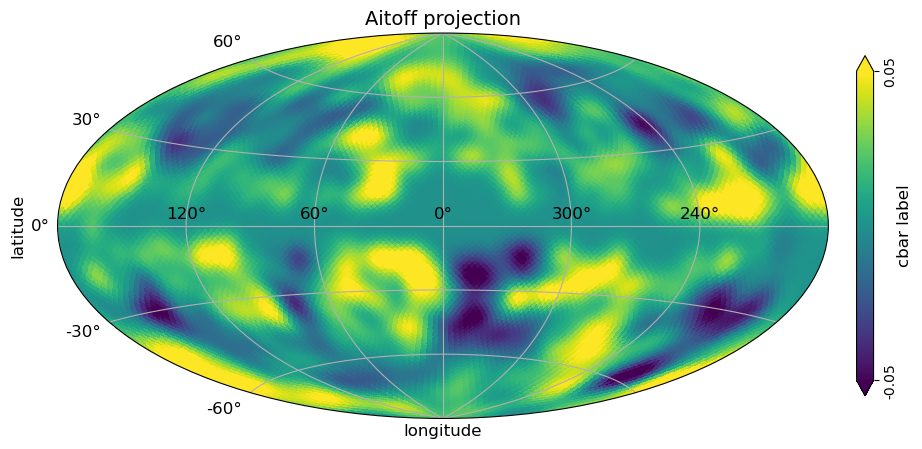
# aitoff view plot with graticule and axis labels and vertical color bar, when min and max is set
# to be different than the min, max of the data, the colorbar is extended
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="aitoff",
title="Aitoff projection",
);
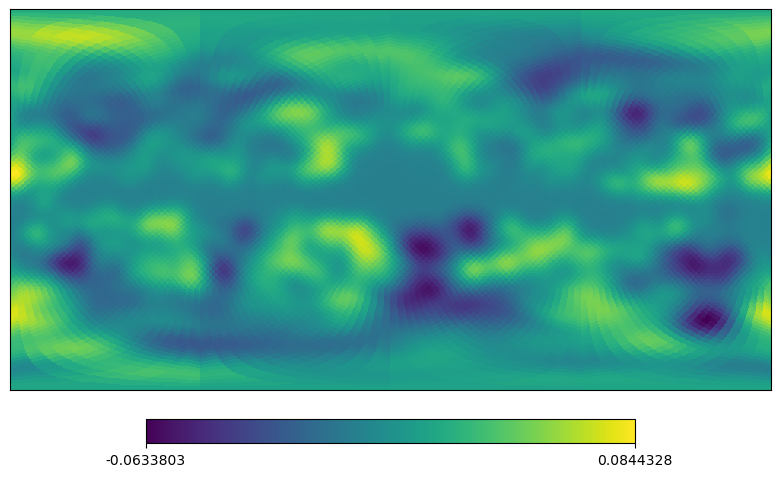
[12]:
# equivalent to cartview in classic healpy
projview(m, coord=["G"], projection_type="cart");
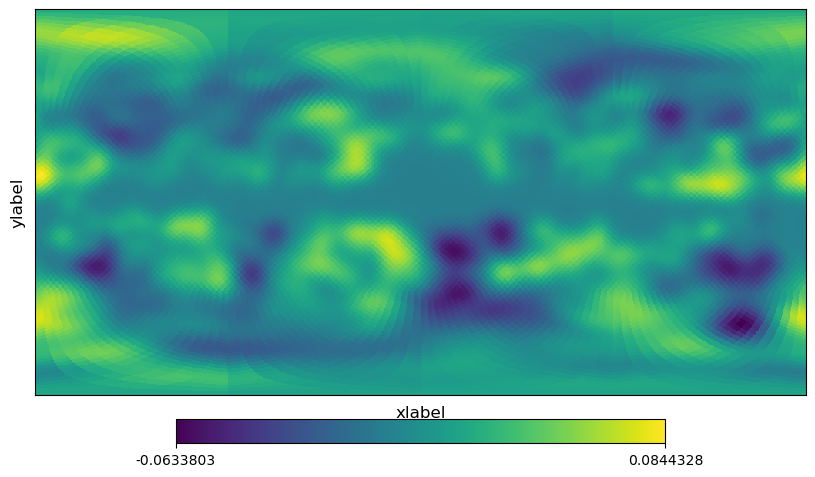
[13]:
# equivalent to cartview
projview(m, coord=["G"], projection_type="cart", xlabel="xlabel", ylabel="ylabel");
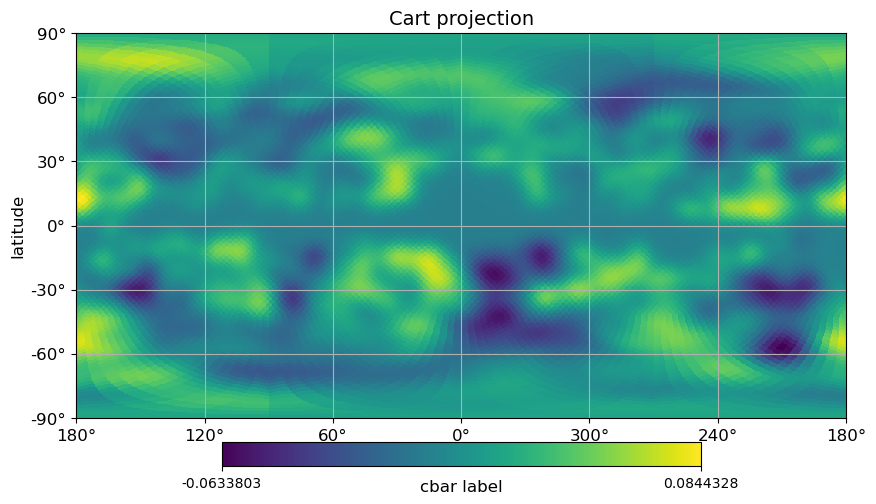
[14]:
# cartview, with labels and graticule
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="horizontal",
projection_type="cart",
title="Cart projection",
);
[15]:
# cartview, with labels and graticule
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="horizontal",
projection_type="cart",
);
[16]:
# cartview, with labels and graticule, vertical cbar
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
projection_type="cart",
);
[17]:
# 3d view
projview(
m,
coord=["G"],
hold=False,
graticule=True,
graticule_labels=True,
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="horizontal",
projection_type="3d",
title="3D projection",
);
[18]:
# 3d view, vertical cbar
projview(
m,
coord=["G"],
hold=False,
graticule=True,
graticule_labels=True,
projection_type="3d",
unit="cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
cmap="viridis",
);
[19]:
# polar view
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
cb_orientation="horizontal",
projection_type="polar",
title="Polar projection",
);
[20]:
# polar view, vertical cbar
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit="cbar label",
cb_orientation="vertical",
projection_type="polar",
);
[21]:
# polar view, override example
projview(
m,
coord=["G"],
hold=False,
graticule=True,
graticule_labels=True,
flip="astro",
projection_type="polar",
unit="cbar label",
cb_orientation="horizontal",
override_plot_properties={
"cbar_shrink": 0.5,
"cbar_pad": 0.02,
"cbar_label_pad": -35,
"figure_width": 16,
"figure_size_ratio": 0.63,
},
);
/home/docs/checkouts/readthedocs.org/user_builds/healpy/conda/latest/lib/python3.9/site-packages/healpy/newvisufunc.py:407: UserWarning:
*** Overriding default plot properies: {'cbar_shrink': 0.4, 'cbar_pad': 0.01, 'cbar_label_pad': 0, 'cbar_tick_direction': 'out', 'vertical_tick_rotation': 90, 'figure_width': 12, 'figure_size_ratio': 0.63} ***
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/healpy/conda/latest/lib/python3.9/site-packages/healpy/newvisufunc.py:411: UserWarning:
*** New plot properies: {'cbar_shrink': 0.5, 'cbar_pad': 0.02, 'cbar_label_pad': -35, 'cbar_tick_direction': 'out', 'vertical_tick_rotation': 90, 'figure_width': 16, 'figure_size_ratio': 0.63} ***
warnings.warn("\n *** New plot properies: " + str(plot_properties) + " ***")
[22]:
# Hammer projection, changed fontsize example & LaTex font test
# and changed color of the axis ticks and graticule
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit=r"cbar label $\alpha$",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="hammer",
title="Hammer projection",
fontsize={
"xlabel": 20,
"ylabel": 20,
"xtick_label": 20,
"ytick_label": 20,
"title": 20,
"cbar_label": 20,
"cbar_tick_label": 20,
},
xtick_label_color="r",
ytick_label_color="g",
graticule_color="black",
);
[23]:
# Hammer projection, no x-axis (phi or longitudinal);; no phi tick label shift
# This shows the true xtick labels
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit=r"cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="hammer",
title="Hammer projection",
phi_convention="symmetrical",
);
# overlay the plot with another data
s = 500
plt.scatter(np.deg2rad(0), np.deg2rad(0), color="r", marker="x", linewidth=10, s=s);
plt.scatter(np.deg2rad(120), np.deg2rad(0), color="r", marker="x", linewidth=10, s=s);
plt.scatter(np.deg2rad(-120), np.deg2rad(0), color="r", marker="x", linewidth=10, s=s);
plt.scatter(np.deg2rad(0), np.deg2rad(60), color="r", marker="x", linewidth=10, s=s);
plt.scatter(np.deg2rad(0), np.deg2rad(-60), color="r", marker="x", linewidth=10, s=s);
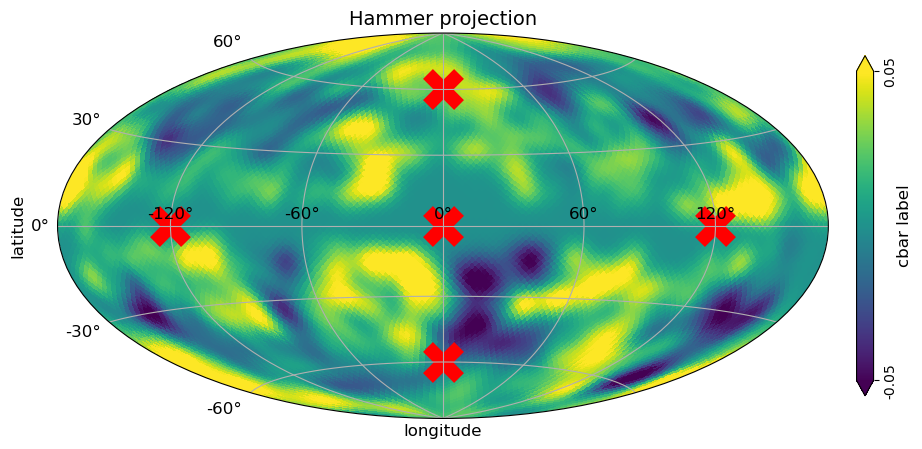
[24]:
# Hammer projection, override axis tick labels
projview(
m,
coord=["G"],
graticule=True,
graticule_labels=True,
unit=r"cbar label",
xlabel="longitude",
ylabel="latitude",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="hammer",
title="Hammer projection",
custom_xtick_labels=["A", "B", "C", "D", "E"],
custom_ytick_labels=["F", "G", "H", "I", "J"],
);
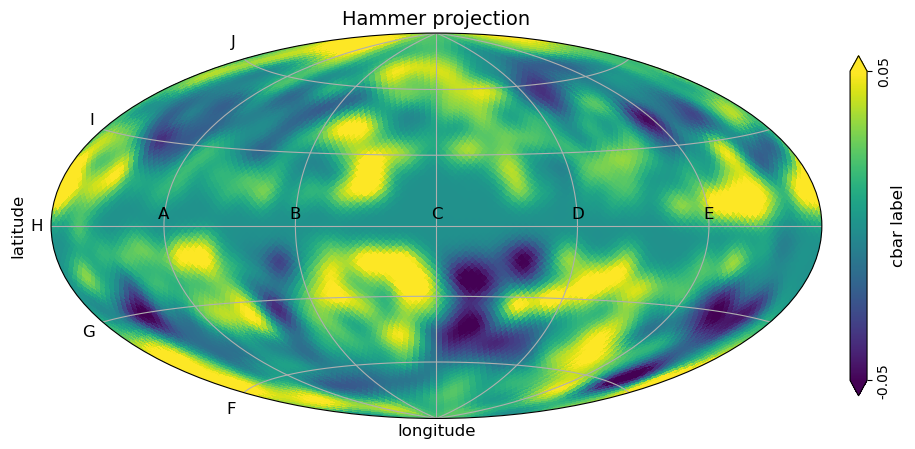
[25]:
# Hammer projection, Equatorial coordinates
projview(
m,
coord=["G", "C"],
graticule=True,
graticule_labels=True,
unit=r"cbar label",
xlabel="RA",
ylabel="DEC",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="hammer",
title="Hammer projection",
);
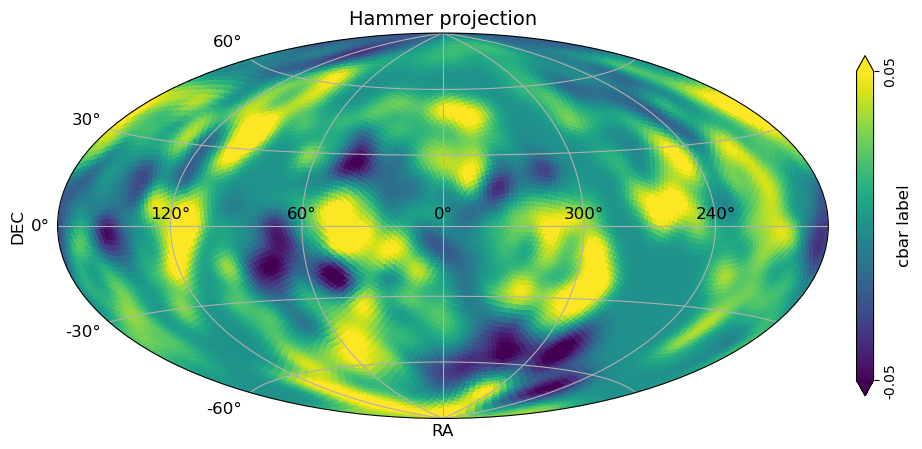
[26]:
# Cartesian projection in Local coordinates (transformed from Galaxy coordinates through Equatorial to local);
# (This is a Galaxy radio emission apex at Pierre Auger Observatory);
# azimuth convention with "phi_convention='counterclockwise'" is North:0, East:270, South:180, West:90
local_sidereal_time = 18
altitude = -35.206667
rotAngles = [(180 + (local_sidereal_time * 15)) % 360, -(altitude - 90)]
projview(
m,
coord=["G", "C"],
graticule=True,
graticule_labels=True,
unit=r"cbar label",
xlabel="azimuth",
ylabel="zenith",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="cart",
title="Cartesian projection",
rot=rotAngles,
fontsize={"xtick_label": 20},
phi_convention="counterclockwise",
);
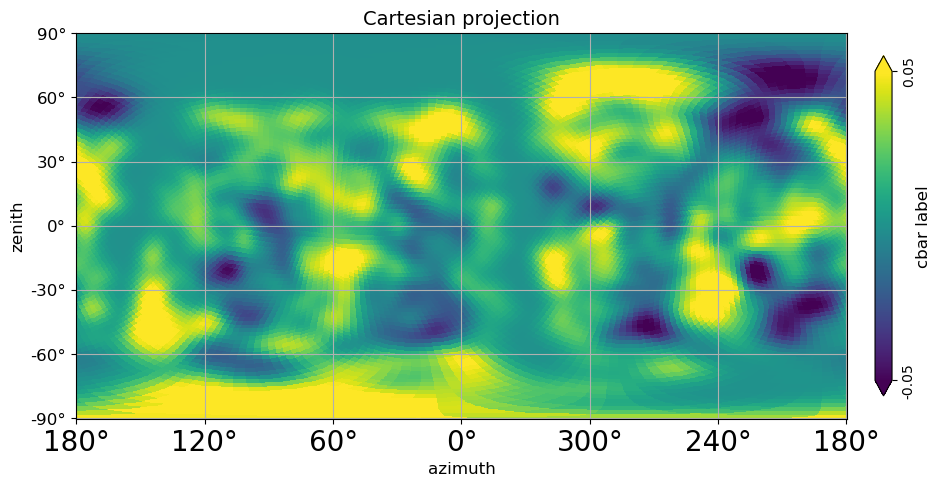
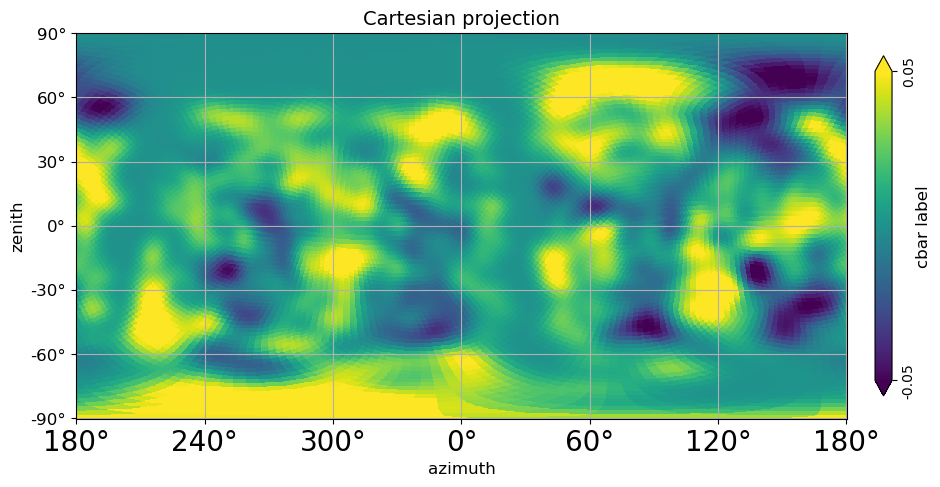
[27]:
# Cartesian projection in Local coordinates (transformed from Galaxy coordinates through Equatorial to local);
# (This is the Galaxy radio emission apex at Pierre Auger Observatory);
# azimuth convention with "phi_convention='clockwise'" is North:0, East:90, South:180, West:270
local_sidereal_time = 18
altitude = -35.206667
rotAngles = [(180 + (local_sidereal_time * 15)) % 360, -(altitude - 90)]
projview(
m,
coord=["G", "C"],
graticule=True,
graticule_labels=True,
unit=r"cbar label",
xlabel="azimuth",
ylabel="zenith",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="cart",
title="Cartesian projection",
rot=rotAngles,
fontsize={"xtick_label": 20},
phi_convention="clockwise",
);
[28]:
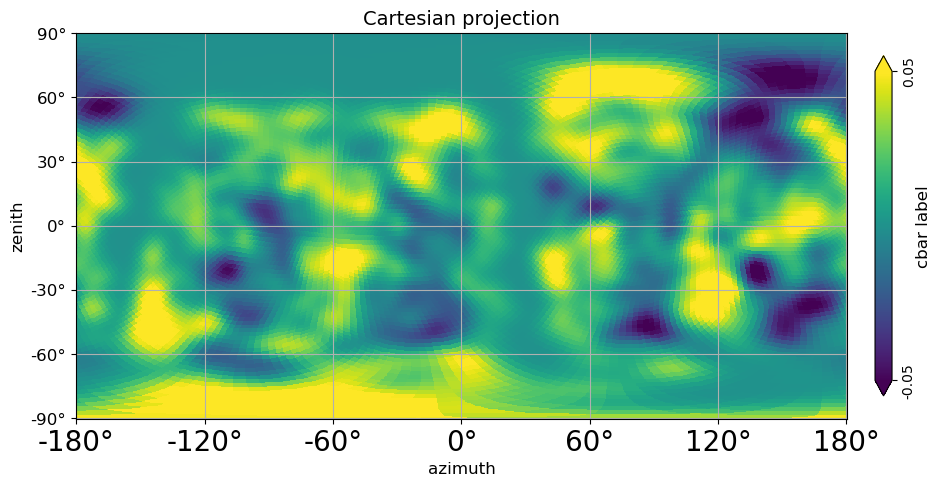
# Cartesian projection in Local coordinates (transformed from Galaxy coordinates through Equatorial to local);
# (This is the Galaxy radio emission apex at Pierre Auger Observatory);
# azimuth convention with "phi_convention='symmetrical'" is North:0, East:90, South:180, West:-90
local_sidereal_time = 18
altitude = -35.206667
rotAngles = [(180 + (local_sidereal_time * 15)) % 360, -(altitude - 90)]
projview(
m,
coord=["G", "C"],
graticule=True,
graticule_labels=True,
unit=r"cbar label",
xlabel="azimuth",
ylabel="zenith",
cb_orientation="vertical",
min=-0.05,
max=0.05,
latitude_grid_spacing=30,
projection_type="cart",
title="Cartesian projection",
rot=rotAngles,
fontsize={"xtick_label": 20},
phi_convention="symmetrical",
)
plt.show()
[29]:
# return only data
# [longitude, latitude, grid_map]
# longitude, latitude are 1D arrays to convert them, to 2D arrays for the plot use np.meshgrid(longitude, latitude);;
# longtitude goes from, -pi to pi (-180 to 180 in degs);
# latitude goes from, -pi/2 to pi/2 (-90 to 90 in degs);
longitude, latitude, grid_map = projview(m, coord=["G"], return_only_data=True)
print(longitude.shape)
print(latitude.shape)
print(grid_map.shape)
(1000,)
(500,)
(500, 1000)
[30]:
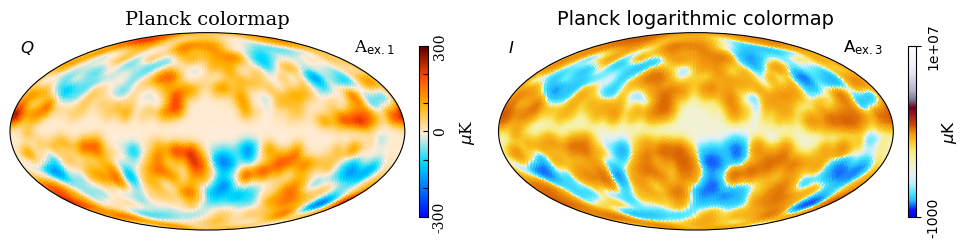
# This example shows two Planck colormaps.
# The planck_log-colormap is used in combination with
# symlog2 for optimal viewing of frequency maps.
#
# Additional displayed features:
# Subfigures (Work the same way as mollview)
# Serif font
# Show tickmarkers
# tick direction keyword
# dipole/monopole subtraction
m_scaled = m*3000
projview(
m_scaled,
title="Planck colormap",
cmap="planck",
rlabel=r"A$_{\mathsf{ex. 1}}$",
llabel=r"$Q$",
unit=r"$\mu$K",
fontname="serif",
width=10,
show_tickmarkers=True,
cbar_ticks=[-300, 0, 300],
cb_orientation="vertical",
sub=121,
override_plot_properties={"cbar_tick_direction": "in"},
);
projview(
m_scaled,
title="Planck logarithmic colormap",
cmap="planck_log",
norm="symlog2",
rlabel=r"A$_{\mathrm{ex. 3}}$",
llabel=r"$I$",
unit=r"$\mu$K",
min=-1e3,
max=1e7,
cb_orientation="vertical",
sub=122,
);
plt.tight_layout();
/home/docs/checkouts/readthedocs.org/user_builds/healpy/conda/latest/lib/python3.9/site-packages/healpy/newvisufunc.py:407: UserWarning:
*** Overriding default plot properies: {'cbar_shrink': 0.6, 'cbar_pad': 0.03, 'cbar_label_pad': 8, 'cbar_tick_direction': 'out', 'vertical_tick_rotation': 90, 'figure_width': 10, 'figure_size_ratio': 0.63} ***
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/healpy/conda/latest/lib/python3.9/site-packages/healpy/newvisufunc.py:411: UserWarning:
*** New plot properies: {'cbar_shrink': 0.6, 'cbar_pad': 0.03, 'cbar_label_pad': 8, 'cbar_tick_direction': 'in', 'vertical_tick_rotation': 90, 'figure_width': 10, 'figure_size_ratio': 0.63} ***
warnings.warn("\n *** New plot properies: " + str(plot_properties) + " ***")
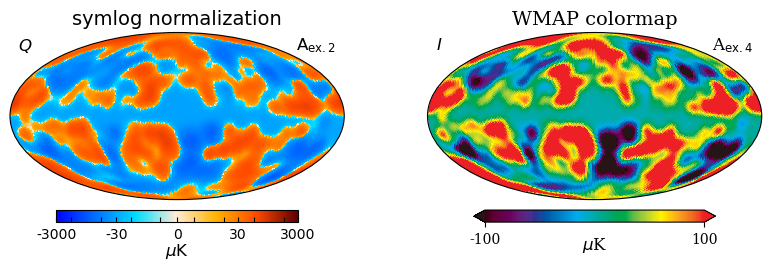
[31]:
# This code shows some of the features listed above,
# in addition to the WMAP colormap.
projview(
m_scaled,
title="symlog normalization",
cmap="planck",
norm="symlog",
rlabel=r"A$_{\mathsf{ex. 2}}$",
llabel=r"$Q$",
unit=r"$\mu$K",
cbar_ticks=[-3000, -30, 0, 30, 3000],
remove_mono=True,
show_tickmarkers=True,
sub=121,
override_plot_properties={"cbar_tick_direction": "in"},
norm_dict={"linscale": 0.5},
);
projview(
m_scaled,
title="WMAP colormap",
cmap="wmap",
rlabel=r"A$_{\mathrm{ex. 4}}$",
llabel=r"$I$",
unit=r"$\mu$K",
fontname="serif",
min=-100,
max=100,
sub=122,
);
plt.tight_layout();
/home/docs/checkouts/readthedocs.org/user_builds/healpy/conda/latest/lib/python3.9/site-packages/healpy/newvisufunc.py:407: UserWarning:
*** Overriding default plot properies: {'cbar_shrink': 0.6, 'cbar_pad': 0.05, 'cbar_label_pad': 0, 'cbar_tick_direction': 'out', 'vertical_tick_rotation': 90, 'figure_width': 8.5, 'figure_size_ratio': 0.63} ***
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/healpy/conda/latest/lib/python3.9/site-packages/healpy/newvisufunc.py:411: UserWarning:
*** New plot properies: {'cbar_shrink': 0.6, 'cbar_pad': 0.05, 'cbar_label_pad': 0, 'cbar_tick_direction': 'in', 'vertical_tick_rotation': 90, 'figure_width': 8.5, 'figure_size_ratio': 0.63} ***
warnings.warn("\n *** New plot properies: " + str(plot_properties) + " ***")